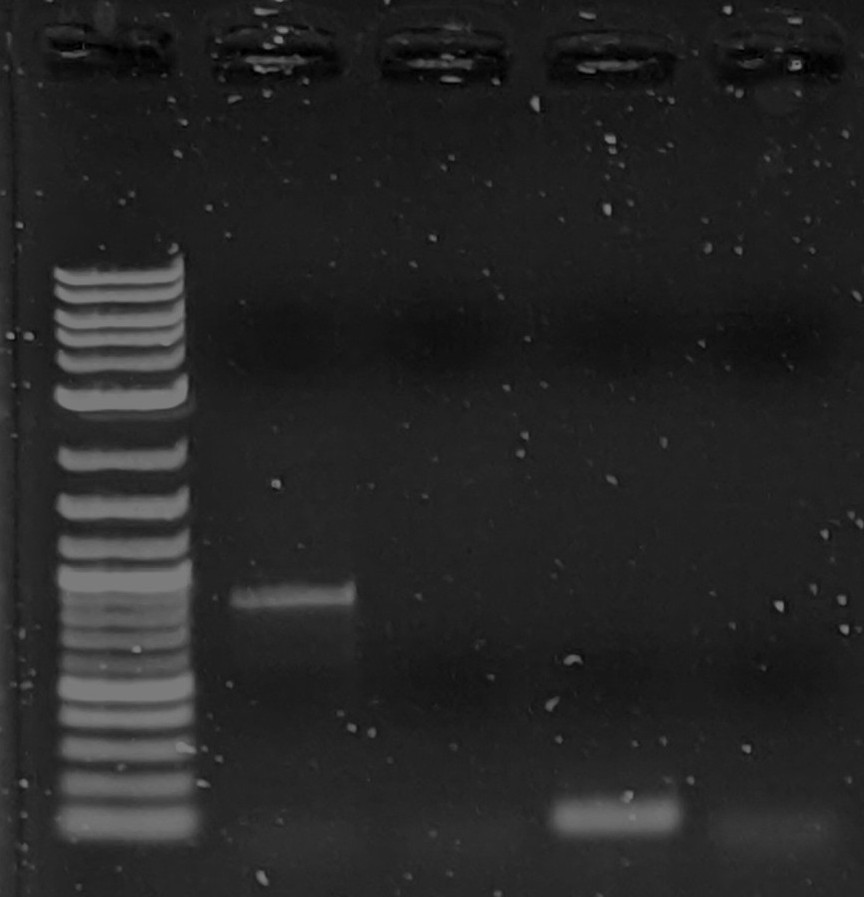
Those low molecular weight bands under 100 bp are almost certainly primer dimers (the curse of any PCR reaction). Primers can combine in funky ways to make products longer than a single primer but basically useless.Since it shows up in the control, I am going to guess the PCR cycle may be part of the issue. Troubleshooting this won't necessarily be quick but the top reasons for primer dimers are:1. Too low annealing temperature or too long annealing time; lower temps amplify less specifically and longer annealing raises the likelihood that non-specific products like primer dimers pop up47C is on the low end for anneal temps so try a degree or two higher to see if this helps. Also look up touchdown PCR which takes a lot of manual work but starts at higher temps for annealing and steadily decreases temp every couple cycles until you hit 47C. Annealing at higher temps helps select more specific products which then have a head start when you hit 47C2. Slow ramp rate on the PCR. Usually not an issue but since you are so low on the annealing temp a slow ramp down from the denaturation and ramp up to the extension can extend the annealing time and cause problem #1Your primer concentration shouldn't be too high at 0.36 uM. If nothing else works sometimes adding additives helps but then you have to get more reagents. DMSO, betaine, BSA, and trehalose are pretty common. I doubt you need to go this far though since you got it to work once already.This page can help: https://www.bio-rad.com/en-us/applications-technologies/pcr-troubleshooting?ID=LUSO3HC4SReggie--On Wednesday, January 26, 2022 at 7:22:42 PM UTC-5 betanic wrote:Thank you for all the suggestions. I got 1 band that appears the correct size after Reggie's suggestion of increasing primer concentration, (I doubled it to 0.36 uM final). Many attempts after that using the same parameters have failed. So I am still unsure what I am doing wrong. I tried increasing the template amount to 5ng and still no bands.Another question: I am getting frequent apparent bands in all samples including negative control right at the end of the range of my ladder (< 100bp). Are these primers showing up? These primers are only 17bp long though...Lanes 3 and 5 are negative control. (pardon the compressed gel, I did a short 23min run) This is using identical parameters (except higher dna concentration) to that of the successful PCR (shown later, below).While I'm at it I will add the 1 successful pcr run as well:Here in Lane 2 is the successful PCR showing to be about 1kb (expected).
Lane 3 was a fail and Lane 4 I suspect was successful while the target sequence was shorter than expected. (my guess)(for reference I am using neb 1kb plus dna ladder)Thanks again for all of your advice.On Mon, Jan 24, 2022 at 5:23 AM Reginald Smith <rsm...@supremevinegar.com> wrote:I second the thermocycler lid temp comments above. Also, I would never use tap water but grocery/drug store distilled water has always worked in my experience. However, I have a few comments/questions on the master mix:1. Why did you add the primers to the master mix? Typically you have a master mix that is primer free so it can be used for multiple types of experiments, especially given how expensive taq polymerase is. No problem now but I would leave the primers out of future master mixes2. The master mix ratio seems quite high though the final concentrations aren't off too much. Typically (though not always) you make a master mix that is a multiple of reaction concentration, for example, 2X so that primers, ddH20, and template together are the same volume as the master mix in the reaction tube. This also allows you to replace some of the ddH20 concentration in the reaction tube with additives like DMSO or trehalose if you need those to get the reaction to work.3. I suspect part of the problem may be the final primer concentration being low. Given the calcs above, your final primer reaction concentration is about 0.18 uM. Typically, you want between 0.2 - 0.5 uM. I would add primer to get up to about 0.4 uM final concentration in the reaction and see if that helps. If it causes problems try different concentrations down to 0.2 uM for each primer.I hope this helps. PCR can be super frustrating sometimes but getting the right band is worth it!Reggie--On Saturday, January 22, 2022 at 2:10:44 AM UTC-5 djwr...@gmail.com wrote:https://www.plasmidsaurus.com/25 dollars to find out if the plasmid is what you think it is.Sent from my iPhoneOn Jan 21, 2022, at 10:05 PM, Koeng <koen...@gmail.com> wrote:> alleged sequenceheh, that is at least what Twist told us it was. I agree with tk though, heated lid could be a big big problem. I always just set it to like 100c.On Friday, January 21, 2022 at 5:15:01 PM UTC-8 betanic wrote:Hello all, I am trying to do my first successful PCR reaction and I am on my 8th attempt, and still have gotten no bands. This is just a control reaction using plasmid templates which should contain the matching primer sequences. Most of the info here is the details of my reagents and at the end is my master mix ingredient list and PCR protocol. Any help would be really appreciated.--- Plasmid Templates:I am using mainly freegenes plasmids, which are mostly biobrick parts and it seems every single one has M13 sequences flanking the genes. Here is an example plasmid with it's alleged sequence (one of the ones I am using as a template): gfasPurple_BB3335I have been miniprepping the plasmids from e.coli dh5a getting 20-100 ng/ul concentrations using neb miniprep kit and eluting with neb monarch elution buffer (which is supposedly TE buffer).--- Primers:First, I ordered "readymade" m13 primers from IDT specifically the "m13 forward (-20)" (GTA AAA CGA CGG CCA GT) and "m13 reverse (-27)" (CAG GAA ACA GCT ATG AC). Both are 17 bp long. They came as a pair of seemingly empty 2ml tubes with 10ug of primers in each tube which I then pipetted 100ul DI h20 into and verified ~100ng/ul using nanodrop. I calculated this to be ~20uM each based on ~5200MW on each primer.--- Taq Kit:I ordered neb taq pcr kit which includes 10x standard taq buffer, 5U/ul Taq DNA Polymerase, and 10mM each dntps solution. The whole kit I have been keeping in a -20c freezer.--- Water SourceThe DI water I am using for my reactions is a cheap filtration kit just like this one. It includes a sediment, carbon, and block filter, reverse osmosis membrane, and mixed bed deionized filter. I read elsewhere that you can do PCR with tap water and so I figured this was sufficient, but I've never done PCR before so I actually have no idea.--- ProcedureOkay onto my procedure. I first diluted my plasmid extractions to ~ 1ng/ul to serve as templates. These should supposedly have m13 flanking sequences, and I add 2ul of this to each reaction tube for a total of 2ng of plasmid DNA. I setup my master mix as below, which is based on neb's taq protocol as a guideline.> 10x standard taq buffer: 20 ul> 10mM DNTPs mix: 4 ul> 19uM m13 fwd primer: 2 ul> 19uM m13 rev primer: 2 ul> 5U/ul taq dna pol: 1 ul> DI H2O: 165 ulTotal 194 ul master mix, which leaves 6ul of volume for template DNA addition. Final reaction vessel mixtures are below, put in standalone 0.2ml PCR tubes.A: 2 ul plasmid1 (~2 ng), + 48 ul master mixB: 2 ul plasmid2 (~2 ng) + 48 ul master mixC: 2 ul plasmid3 (~2 ng) + 48 ul master mixD: 50 ul master mix negative controlThe thermal cycler I am using is a MJ-Research PTC-200 "DNA Engine" with Dual Block head. I set the protocol to use a constant temp heated lid to 60c and temp control method to "calculated" based on 50ul reaction in PCR tubes. The program was set to the following:1: 95c 1min2: 95c 30s3: 47c 30s4: 68c 1min5: loop 2-4 29x6: 68c 5min7: 10c holdThe anneal temperature of 47c was chosen based off of neb's Tm calculator and given the m13 primer sequences and final primer concentration of 200nM. The extension time is based off of neb's pcr protocol guideline of 1min per kb of target segment length, the target sequences from the plasmid maps for these plasmids are around 700-1000 bp.--- Final Notes:There was considerable condensation near the lid of the tubes at the end of my last reaction, I don't know how much of an affect this would have.I have altered the above protocol a couple of times trying 50 and 55c anneal temp and 40s anneal time. I have tried a second taq kit in case my first one had a bad reagent. So far I have not used a different set of primers so that is my next thing to try.I believe I have given all the info I can think of. At this point I have no idea why I am not seeing bands and not sure what to try next. Once again any help or advice would be very appreciated.---- You received this message because you are subscribed to the Google Groups DIYbio group. To post to this group, send email to diy...@googlegroups.com. To unsubscribe from this group, send email to diybio+un...@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/diybio?hl=en
Learn more at www.diybio.org
---
You received this message because you are subscribed to the Google Groups "DIYbio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to diybio+un...@googlegroups.com.To view this discussion on the web visit https://groups.google.com/d/msgid/diybio/65636589-d03b-4384-87f1-6a4edc7c900fn%40googlegroups.com.
-- You received this message because you are subscribed to the Google Groups DIYbio group. To post to this group, send email to diy...@googlegroups.com. To unsubscribe from this group, send email to diybio+un...@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/diybio?hl=en
Learn more at www.diybio.org
---
You received this message because you are subscribed to the Google Groups "DIYbio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to diybio+un...@googlegroups.com.To view this discussion on the web visit https://groups.google.com/d/msgid/diybio/7b5fc2f7-9549-4232-adeb-ac04fb3ce791n%40googlegroups.com.
-- You received this message because you are subscribed to the Google Groups DIYbio group. To post to this group, send email to diybio@googlegroups.com. To unsubscribe from this group, send email to diybio+unsubscribe@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/diybio?hl=en
Learn more at www.diybio.org
---
You received this message because you are subscribed to the Google Groups "DIYbio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to diybio+unsubscribe@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/diybio/5835f78e-d191-4ea2-887e-9d2911c8be22n%40googlegroups.com.
-- You received this message because you are subscribed to the Google Groups DIYbio group. To post to this group, send email to diybio@googlegroups.com. To unsubscribe from this group, send email to diybio+unsubscribe@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/diybio?hl=en
Learn more at www.diybio.org
---
You received this message because you are subscribed to the Google Groups "DIYbio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to diybio+unsubscribe@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/diybio/CALN4Cx2X0kAQHe4hnjtVojvyhY2MW_iKP8M3f3hSPiDDDfB8xA%40mail.gmail.com.